Perform Variance Partition analysis for each assay
Usage
fitVarPart(
x,
formula,
data = colData(x),
assays = assayNames(x),
quiet = FALSE,
BPPARAM = SerialParam(),
...
)
# S4 method for class 'dreamletProcessedData'
fitVarPart(
x,
formula,
data = colData(x),
assays = assayNames(x),
quiet = FALSE,
BPPARAM = SerialParam(),
...
)Arguments
- x
SingleCellExperiment or dreamletProcessedData object
- formula
regression formula for differential expression analysis
- data
metadata used in regression formula
- assays
array of assay names to include in analysis. Defaults to
assayNames(x)- quiet
show messages
- BPPARAM
parameters for parallel evaluation
- ...
other arguments passed to
dream
Value
Object of class vpDF inheriting from DataFrame storing the variance fractions for each gene and cell type.
Examples
library(muscat)
library(SingleCellExperiment)
data(example_sce)
# create pseudobulk for each sample and cell cluster
pb <- aggregateToPseudoBulk(example_sce,
assay = "counts",
cluster_id = "cluster_id",
sample_id = "sample_id",
verbose = FALSE
)
# voom-style normalization
res.proc <- processAssays(pb, ~group_id)
#> B cells...
#> 0.052 secs
#> CD14+ Monocytes...
#> 0.085 secs
#> CD4 T cells...
#> 0.062 secs
#> CD8 T cells...
#> 0.037 secs
#> FCGR3A+ Monocytes...
#> 0.08 secs
# variance partitioning analysis
vp <- fitVarPart(res.proc, ~group_id)
#> B cells...
#> 0.63 secs
#> CD14+ Monocytes...
#> 0.83 secs
#> CD4 T cells...
#> 0.63 secs
#> CD8 T cells...
#> 0.38 secs
#> FCGR3A+ Monocytes...
#> 0.76 secs
#>
# Show variance fractions at the gene-level for each cell type
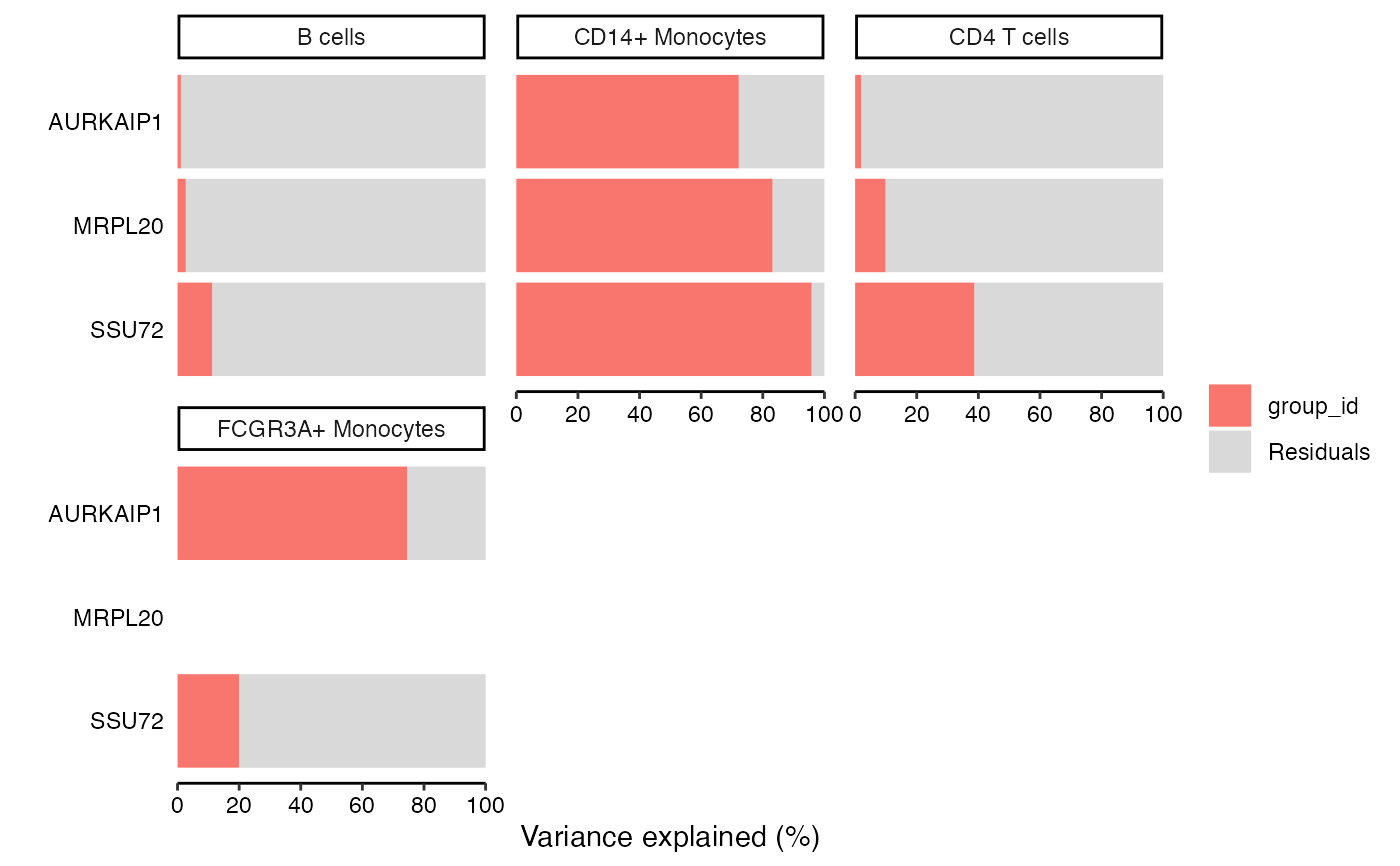
genes <- vp$gene[2:4]
plotPercentBars(vp[vp$gene %in% genes, ])
#> Warning: Ignoring empty aesthetic: `width`.
 # Summarize variance fractions genome-wide for each cell type
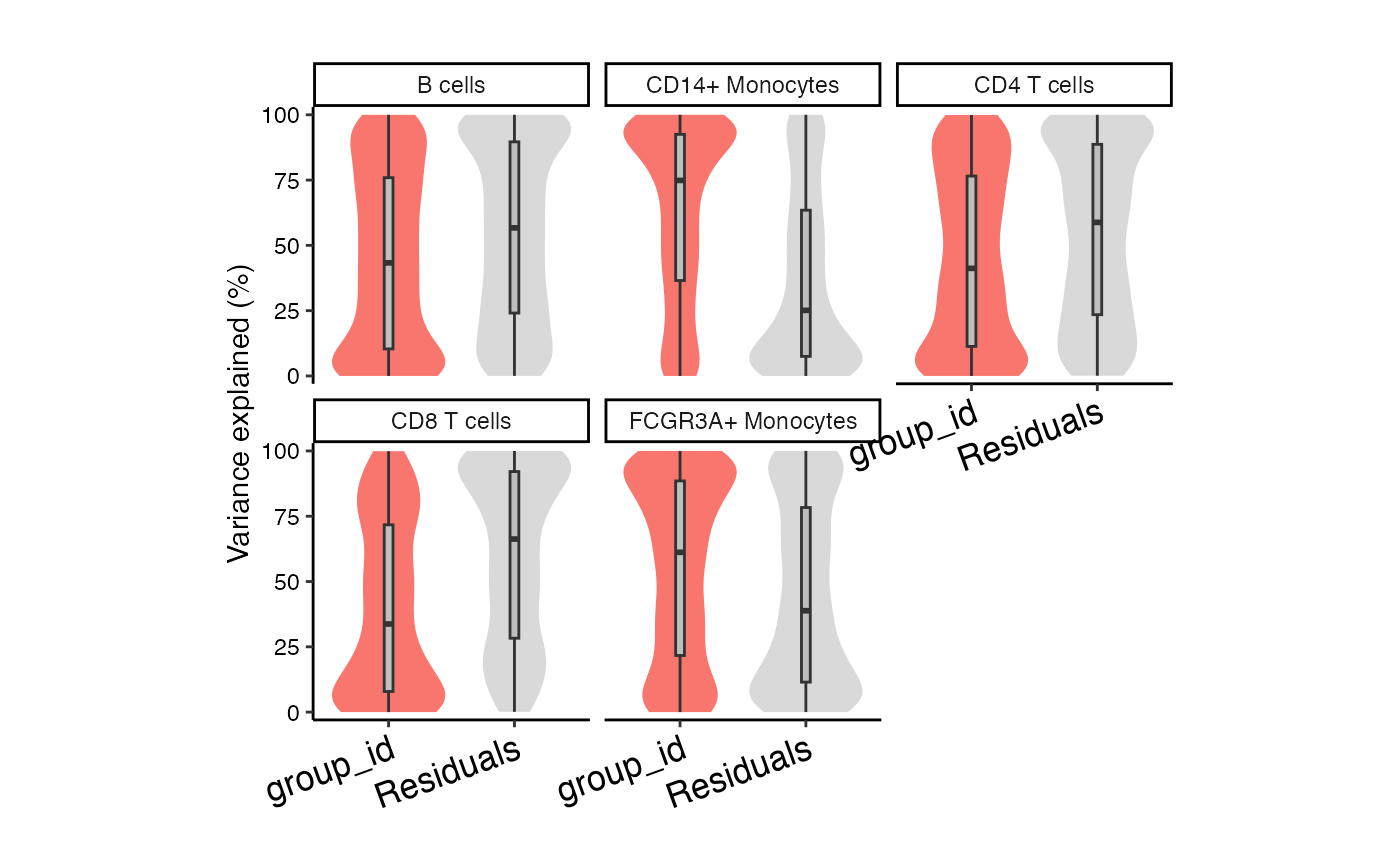
plotVarPart(vp)
# Summarize variance fractions genome-wide for each cell type
plotVarPart(vp)