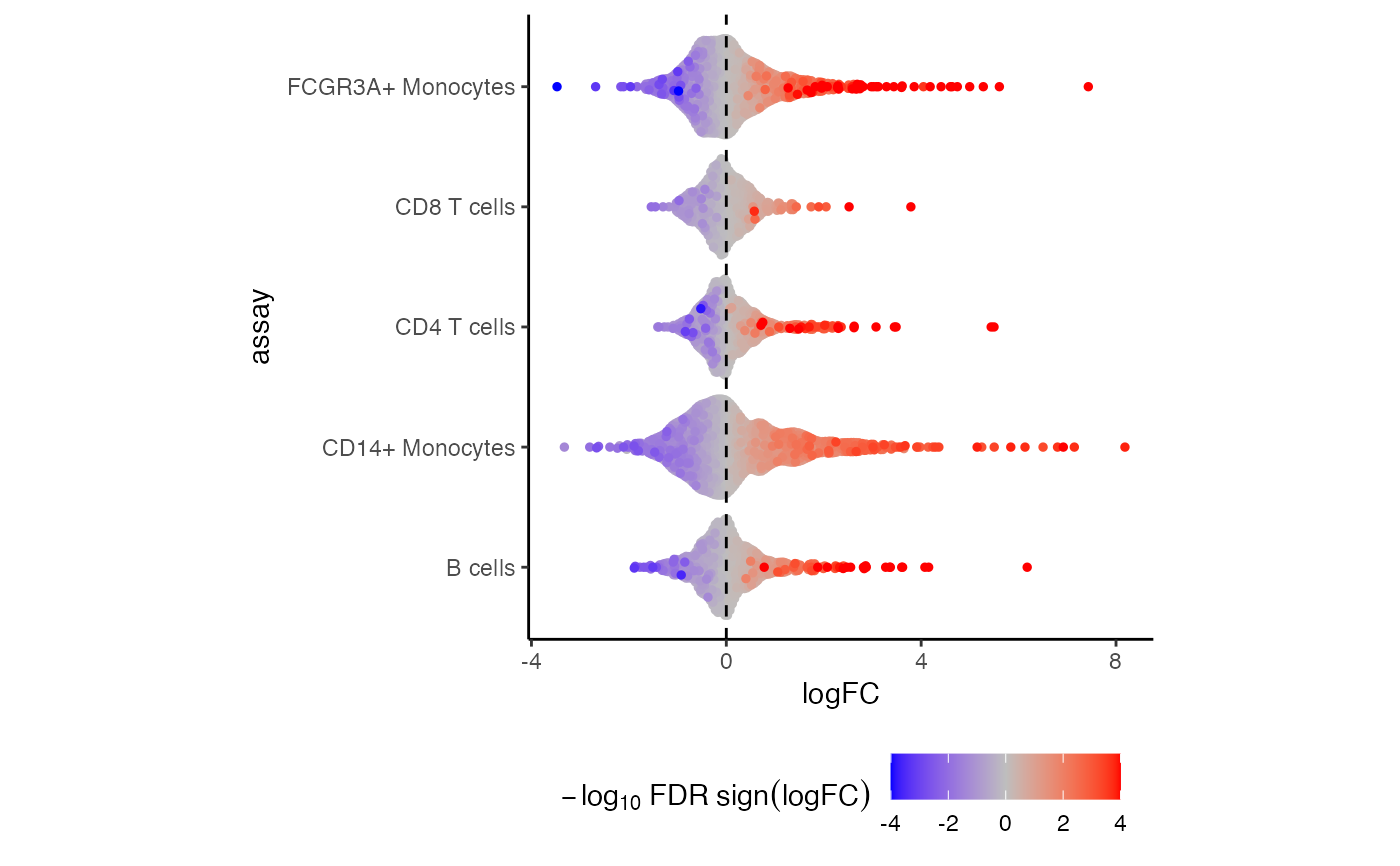
Beeswarm plot of effect sizes for each assay, colored by sign and FDR
Usage
plotBeeswarm(res.dl, coef, fdr.range = 4, assays = assayNames(res.dl))Arguments
- res.dl
dreamletResultobject fromdreamlet()- coef
coefficient name fed to
topTable()- fdr.range
range for coloring FDR
- assays
which assays to plot
Examples
library(muscat)
library(SingleCellExperiment)
data(example_sce)
# create pseudobulk for each sample and cell cluster
pb <- aggregateToPseudoBulk(example_sce,
assay = "counts",
cluster_id = "cluster_id",
sample_id = "sample_id",
verbose = FALSE
)
# voom-style normalization
res.proc <- processAssays(pb, ~group_id)
#> B cells...
#> 0.053 secs
#> CD14+ Monocytes...
#> 0.084 secs
#> CD4 T cells...
#> 0.061 secs
#> CD8 T cells...
#> 0.42 secs
#> FCGR3A+ Monocytes...
#> 0.079 secs
# Differential expression analysis within each assay,
# evaluated on the voom normalized data
res.dl <- dreamlet(res.proc, ~group_id)
#> B cells...
#> 0.048 secs
#> CD14+ Monocytes...
#> 0.063 secs
#> CD4 T cells...
#> 0.051 secs
#> CD8 T cells...
#> 0.034 secs
#> FCGR3A+ Monocytes...
#> 0.071 secs
# Beeswarm plot of effect sizes for each assay,
# colored by sign and FDR
plotBeeswarm(res.dl, "group_idstim")