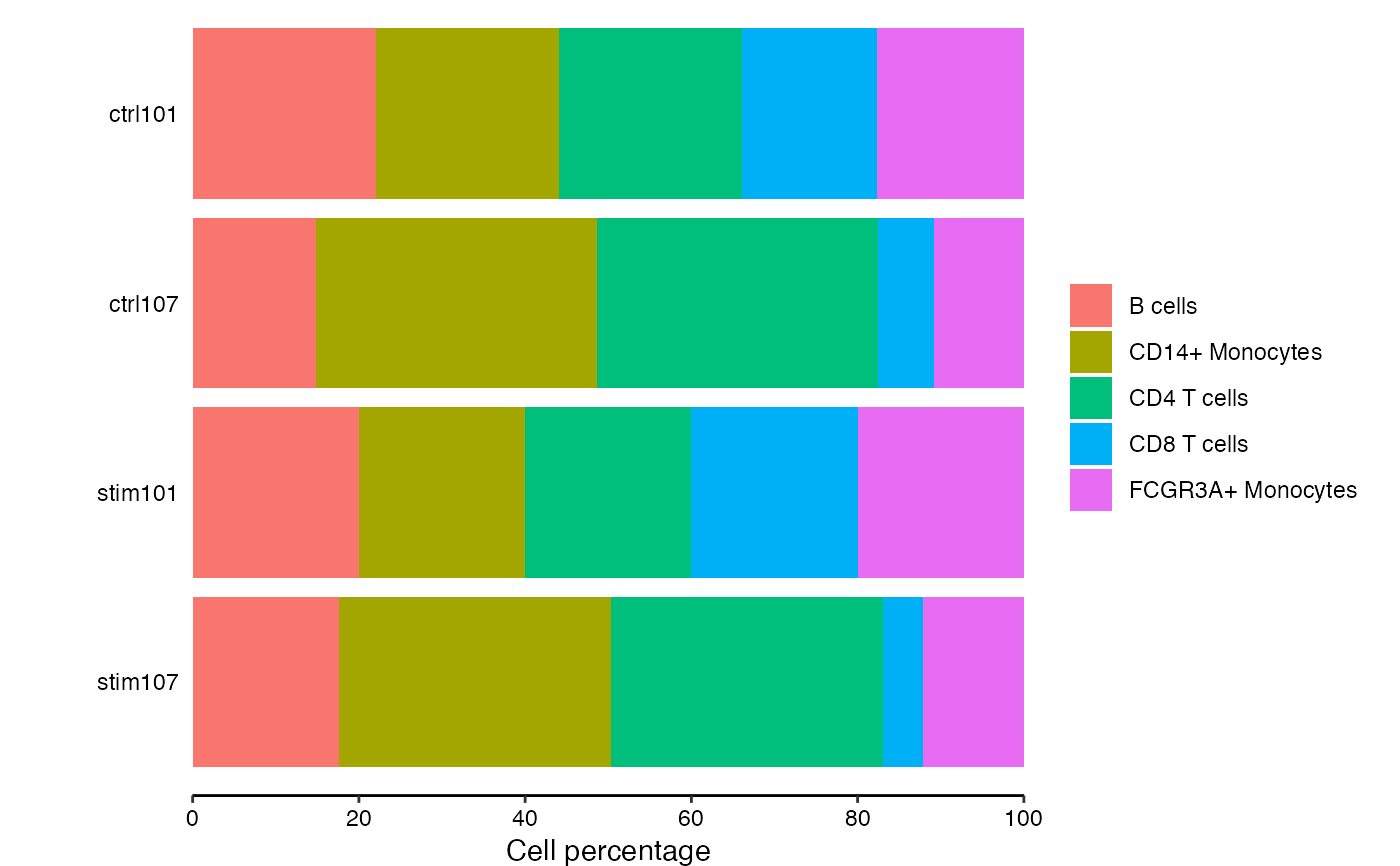
Bar plot of cell compositions
Usage
plotCellComposition(obj, col, width = NULL)
# S4 method for class 'SingleCellExperiment'
plotCellComposition(obj, col, width = NULL)
# S4 method for class 'matrix'
plotCellComposition(obj, col, width = NULL)
# S4 method for class 'data.frame'
plotCellComposition(obj, col, width = NULL)Examples
library(muscat)
library(SingleCellExperiment)
data(example_sce)
# create pseudobulk for each sample and cell cluster
pb <- aggregateToPseudoBulk(example_sce,
assay = "counts",
cluster_id = "cluster_id",
sample_id = "sample_id",
verbose = FALSE
)
# show cell composition bar plots
plotCellComposition(pb)
#> Warning: Ignoring empty aesthetic: `width`.
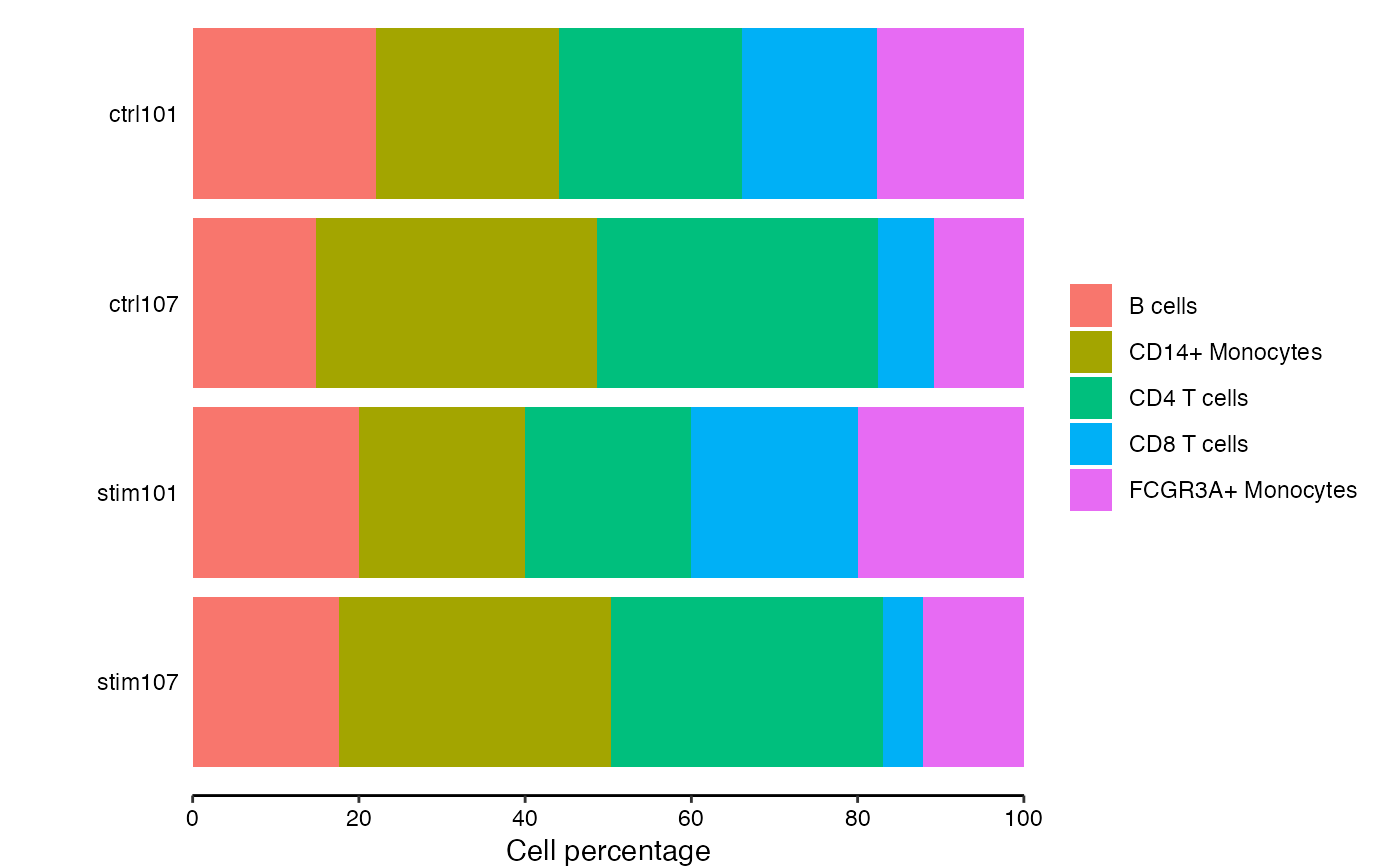 # extract cell counts
df_cellCounts <- cellCounts(pb)
# show cell composition bar plots
plotCellComposition(df_cellCounts)
#> Warning: Ignoring empty aesthetic: `width`.
# extract cell counts
df_cellCounts <- cellCounts(pb)
# show cell composition bar plots
plotCellComposition(df_cellCounts)
#> Warning: Ignoring empty aesthetic: `width`.