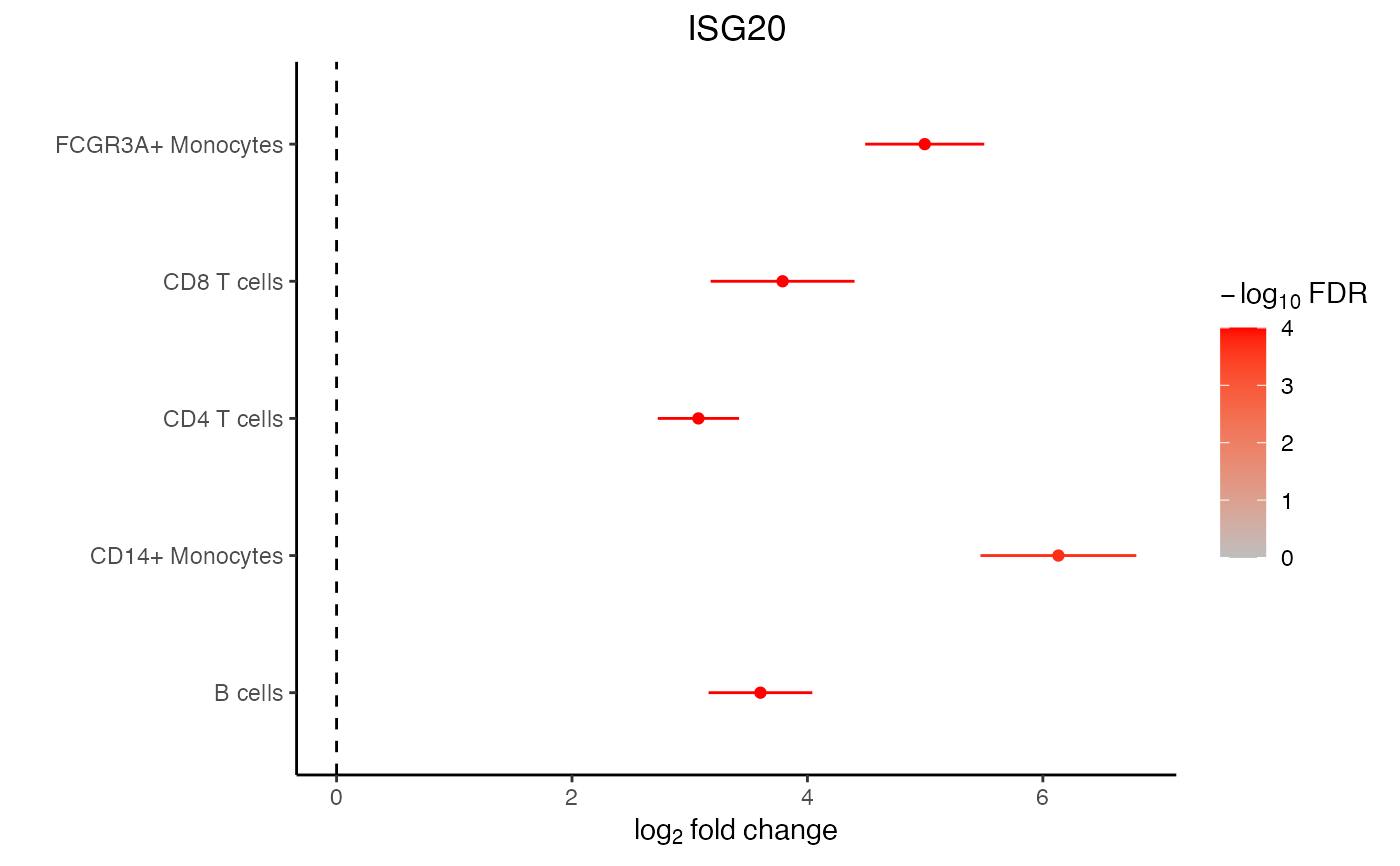
Forest plot
Examples
library(muscat)
library(SingleCellExperiment)
data(example_sce)
# create pseudobulk for each sample and cell cluster
pb <- aggregateToPseudoBulk(example_sce,
assay = "counts",
cluster_id = "cluster_id",
sample_id = "sample_id",
verbose = FALSE
)
# voom-style normalization
res.proc <- processAssays(pb, ~group_id)
#> B cells...
#> 0.06 secs
#> CD14+ Monocytes...
#> 0.084 secs
#> CD4 T cells...
#> 0.061 secs
#> CD8 T cells...
#> 0.037 secs
#> FCGR3A+ Monocytes...
#> 0.079 secs
# Differential expression analysis within each assay,
# evaluated on the voom normalized data
res.dl <- dreamlet(res.proc, ~group_id)
#> B cells...
#> 0.048 secs
#> CD14+ Monocytes...
#> 0.064 secs
#> CD4 T cells...
#> 0.059 secs
#> CD8 T cells...
#> 0.032 secs
#> FCGR3A+ Monocytes...
#> 0.06 secs
# show coefficients estimated for each cell type
coefNames(res.dl)
#> [1] "(Intercept)" "group_idstim"
# Show estimated log fold change with in each cell type
plotForest(res.dl, gene = "ISG20", coef = "group_idstim")