
Integration with dreamlet / SingleCellExperiment
Developed by Gabriel Hoffman
Run on 2025-08-11 14:19:51.385562
Source:vignettes/integration.Rmd
integration.RmdLoad and process single cell data
Here we perform analysis of PBMCs from 8 individuals stimulated with
interferon-β Kang, et
al, 2018, Nature Biotech. We perform standard processing with dreamlet
to compute pseudobulk before applying crumblr.
Here, single cell RNA-seq data is downloaded from ExperimentHub.
library(dreamlet)
library(muscat)
library(ExperimentHub)
library(scater)
# Download data, specifying EH2259 for the Kang, et al. study
eh <- ExperimentHub()
sce <- eh[["EH2259"]]
sce$ind <- as.character(sce$ind)
# only keep singlet cells with sufficient reads
sce <- sce[rowSums(counts(sce) > 0) > 0, ]
sce <- sce[, colData(sce)$multiplets == "singlet"]
# compute QC metrics
qc <- perCellQCMetrics(sce)
# remove cells with few or many detected genes
ol <- isOutlier(metric = qc$detected, nmads = 2, log = TRUE)
sce <- sce[, !ol]
# set variable indicating stimulated (stim) or control (ctrl)
sce$StimStatus <- sce$stimAggregate to pseudobulk
Dreamlet creates the pseudobulk dataset:
# Since 'ind' is the individual and 'StimStatus' is the stimulus status,
# create unique identifier for each sample
sce$id <- paste0(sce$StimStatus, sce$ind)
# Create pseudobulk data by specifying cluster_id and sample_id for aggregating cells
pb <- aggregateToPseudoBulk(sce,
assay = "counts",
cluster_id = "cell",
sample_id = "id",
verbose = FALSE
)Process data
Here we evaluate whether the observed cell proportions change in response to interferon-β.
library(crumblr)
# use dreamlet::cellCounts() to extract data
cellCounts(pb)[1:3, 1:3]## B cells CD14+ Monocytes CD4 T cells
## ctrl101 101 136 288
## ctrl1015 424 644 819
## ctrl1016 119 315 413
# Apply crumblr transformation
# cobj is an EList object compatable with limma workflow
# cobj$E stores transformed values
# cobj$weights stores precision weights
cobj <- crumblr(cellCounts(pb))Analysis
Now continue on with the downstream analysis
library(variancePartition)
fit <- dream(cobj, ~ StimStatus + ind, colData(pb))
fit <- eBayes(fit)
topTable(fit, coef = "StimStatusstim", number = Inf)## logFC AveExpr t P.Value adj.P.Val B
## CD8 T cells -0.25085170 0.0857175 -4.0787416 0.002436375 0.01949100 -1.279815
## Dendritic cells 0.37386979 -2.1849234 3.1619195 0.010692544 0.02738587 -2.638507
## CD14+ Monocytes -0.10525402 1.2698117 -3.1226341 0.011413912 0.02738587 -2.709377
## B cells -0.10478652 0.5516882 -3.0134349 0.013692935 0.02738587 -2.940542
## CD4 T cells -0.07840101 2.0201947 -2.2318104 0.050869691 0.08139151 -4.128069
## FCGR3A+ Monocytes 0.07425165 -0.2567492 1.6647681 0.128337022 0.17111603 -4.935304
## NK cells 0.10270672 0.3797777 1.5181860 0.161321761 0.18436773 -5.247806
## Megakaryocytes 0.01377768 -1.8655172 0.1555131 0.879651456 0.87965146 -6.198336Given the results here, we see that CD8 T cells at others change relative abundance following treatment with interferon-β.
Multivariate testing along a tree
ere we construct a hierarchical clustering between cell types based
on gene expression from pseudobulk, and perform a multivariate test for
each internal node of the tree based on its leaf nodes. The results for
the leaves are the same as from topTable() above.
# hierarchical cluster based on pseudobulked gene expression
hcl <- buildClusterTreeFromPB(pb)
# Perform multivariate test across the hierarchy
res <- treeTest(fit, cobj, hcl, coef = "StimStatusstim")
# Plot hierarchy and testing results
plotTreeTest(res)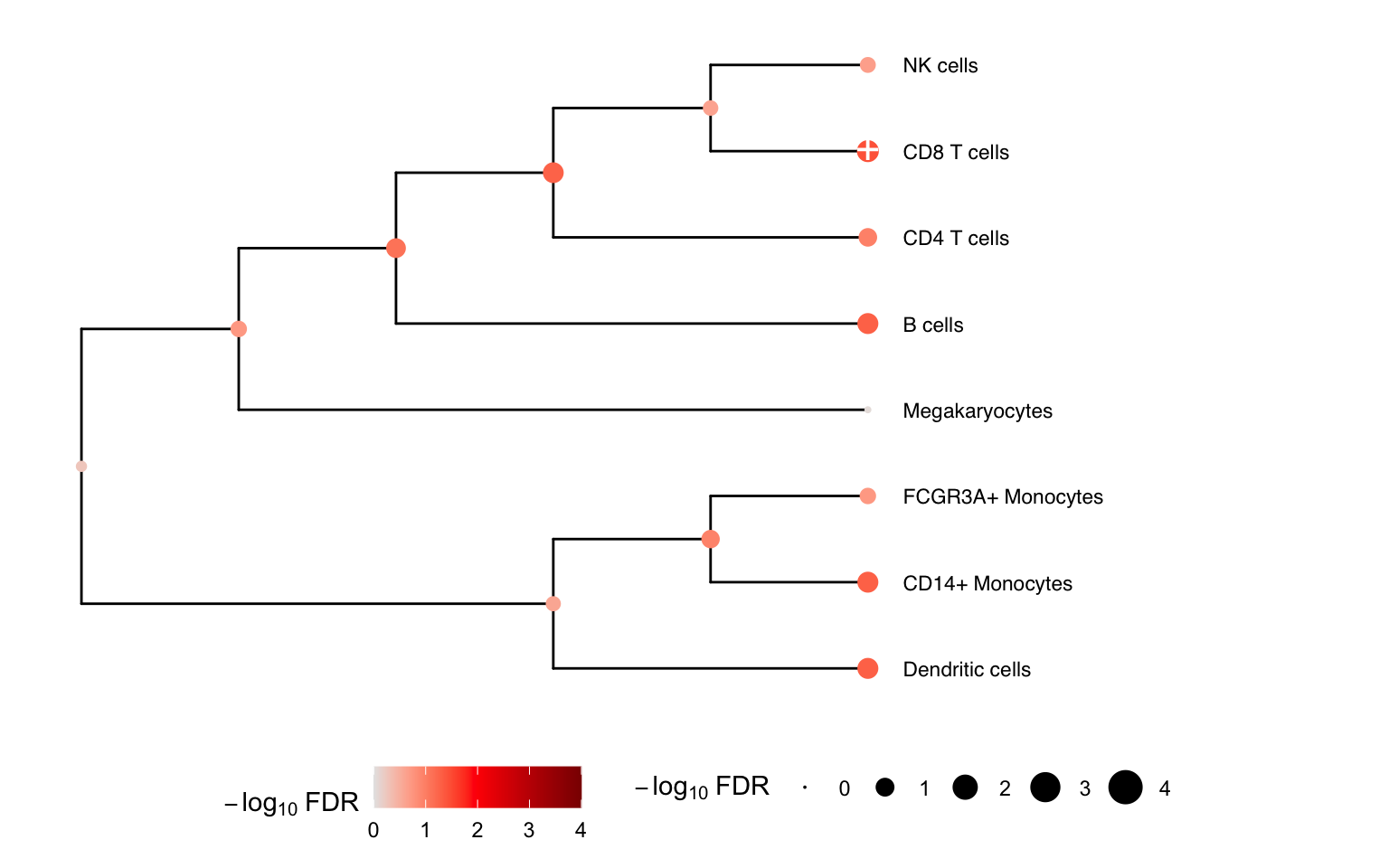
Session Info
## R version 4.5.1 (2025-06-13)
## Platform: aarch64-apple-darwin23.6.0
## Running under: macOS Sonoma 14.7.1
##
## Matrix products: default
## BLAS/LAPACK: /opt/homebrew/Cellar/openblas/0.3.30/lib/libopenblasp-r0.3.30.dylib; LAPACK version 3.12.0
##
## locale:
## [1] en_US.UTF-8/en_US.UTF-8/en_US.UTF-8/C/en_US.UTF-8/en_US.UTF-8
##
## time zone: America/New_York
## tzcode source: internal
##
## attached base packages:
## [1] stats4 stats graphics grDevices utils datasets methods base
##
## other attached packages:
## [1] crumblr_0.99.22 muscData_1.22.0 scater_1.36.0
## [4] scuttle_1.18.0 ExperimentHub_2.16.1 AnnotationHub_3.16.1
## [7] BiocFileCache_2.16.1 dbplyr_2.5.0 muscat_1.22.0
## [10] dreamlet_1.6.0 SingleCellExperiment_1.30.1 SummarizedExperiment_1.38.1
## [13] Biobase_2.68.0 GenomicRanges_1.60.0 GenomeInfoDb_1.44.1
## [16] IRanges_2.42.0 S4Vectors_0.46.0 BiocGenerics_0.54.0
## [19] generics_0.1.4 MatrixGenerics_1.20.0 matrixStats_1.5.0
## [22] variancePartition_1.37.4 BiocParallel_1.42.1 limma_3.64.3
## [25] ggplot2_3.5.2 BiocStyle_2.36.0
##
## loaded via a namespace (and not attached):
## [1] fs_1.6.6 bitops_1.0-9 httr_1.4.7
## [4] RColorBrewer_1.1-3 doParallel_1.0.17 Rgraphviz_2.52.0
## [7] numDeriv_2016.8-1.1 tools_4.5.1 sctransform_0.4.2
## [10] backports_1.5.0 R6_2.6.1 metafor_4.8-0
## [13] lazyeval_0.2.2 mgcv_1.9-3 GetoptLong_1.0.5
## [16] withr_3.0.2 prettyunits_1.2.0 gridExtra_2.3
## [19] cli_3.6.5 textshaping_1.0.1 labeling_0.4.3
## [22] sass_0.4.10 KEGGgraph_1.68.0 SQUAREM_2021.1
## [25] mvtnorm_1.3-3 blme_1.0-6 pkgdown_2.1.3
## [28] mixsqp_0.3-54 yulab.utils_0.2.0 systemfonts_1.2.3
## [31] zenith_1.10.0 parallelly_1.45.1 invgamma_1.2
## [34] RSQLite_2.4.2 gridGraphics_0.5-1 shape_1.4.6.1
## [37] gtools_3.9.5 dplyr_1.1.4 Matrix_1.7-3
## [40] metadat_1.4-0 ggbeeswarm_0.7.2 abind_1.4-8
## [43] lifecycle_1.0.4 yaml_2.3.10 edgeR_4.6.3
## [46] mathjaxr_1.8-0 gplots_3.2.0 SparseArray_1.8.1
## [49] grid_4.5.1 blob_1.2.4 crayon_1.5.3
## [52] lattice_0.22-7 beachmat_2.24.0 msigdbr_25.1.1
## [55] annotate_1.86.1 KEGGREST_1.48.1 pillar_1.11.0
## [58] knitr_1.50 ComplexHeatmap_2.24.1 rjson_0.2.23
## [61] boot_1.3-31 corpcor_1.6.10 future.apply_1.20.0
## [64] codetools_0.2-20 glue_1.8.0 ggfun_0.2.0
## [67] data.table_1.17.8 treeio_1.32.0 vctrs_0.6.5
## [70] png_0.1-8 Rdpack_2.6.4 gtable_0.3.6
## [73] assertthat_0.2.1 cachem_1.1.0 zigg_0.0.2
## [76] xfun_0.52 mime_0.13 rbibutils_2.3
## [79] S4Arrays_1.8.1 Rfast_2.1.5.1 reformulas_0.4.1
## [82] iterators_1.0.14 statmod_1.5.0 nlme_3.1-168
## [85] pbkrtest_0.5.5 ggtree_3.16.3 bit64_4.6.0-1
## [88] filelock_1.0.3 progress_1.2.3 EnvStats_3.1.0
## [91] bslib_0.9.0 TMB_1.9.17 irlba_2.3.5.1
## [94] vipor_0.4.7 KernSmooth_2.23-26 colorspace_2.1-1
## [97] rmeta_3.0 DBI_1.2.3 DESeq2_1.48.1
## [100] tidyselect_1.2.1 bit_4.6.0 compiler_4.5.1
## [103] curl_6.4.0 graph_1.86.0 BiocNeighbors_2.2.0
## [106] desc_1.4.3 DelayedArray_0.34.1 bookdown_0.43
## [109] scales_1.4.0 caTools_1.18.3 remaCor_0.0.18
## [112] rappdirs_0.3.3 stringr_1.5.1 digest_0.6.37
## [115] minqa_1.2.8 rmarkdown_2.29 aod_1.3.3
## [118] XVector_0.48.0 RhpcBLASctl_0.23-42 htmltools_0.5.8.1
## [121] pkgconfig_2.0.3 lme4_1.1-37 sparseMatrixStats_1.20.0
## [124] mashr_0.2.79 fastmap_1.2.0 rlang_1.1.6
## [127] GlobalOptions_0.1.2 htmlwidgets_1.6.4 UCSC.utils_1.4.0
## [130] DelayedMatrixStats_1.30.0 farver_2.1.2 jquerylib_0.1.4
## [133] jsonlite_2.0.0 BiocSingular_1.24.0 RCurl_1.98-1.17
## [136] magrittr_2.0.3 ggplotify_0.1.2 GenomeInfoDbData_1.2.14
## [139] patchwork_1.3.1 Rcpp_1.1.0 ape_5.8-1
## [142] babelgene_22.9 viridis_0.6.5 EnrichmentBrowser_2.38.0
## [145] stringi_1.8.7 MASS_7.3-65 plyr_1.8.9
## [148] listenv_0.9.1 parallel_4.5.1 ggrepel_0.9.6
## [151] Biostrings_2.76.0 splines_4.5.1 hms_1.1.3
## [154] circlize_0.4.16 locfit_1.5-9.12 reshape2_1.4.4
## [157] ScaledMatrix_1.16.0 BiocVersion_3.21.1 XML_3.99-0.18
## [160] evaluate_1.0.4 RcppParallel_5.1.10.9000 BiocManager_1.30.26
## [163] nloptr_2.2.1 foreach_1.5.2 tidyr_1.3.1
## [166] purrr_1.1.0 future_1.67.0 clue_0.3-66
## [169] scattermore_1.2 ashr_2.2-63 rsvd_1.0.5
## [172] broom_1.0.9 xtable_1.8-4 tidytree_0.4.6
## [175] fANCOVA_0.6-1 viridisLite_0.4.2 ragg_1.4.0
## [178] truncnorm_1.0-9 tibble_3.3.0 aplot_0.2.8
## [181] lmerTest_3.1-3 glmmTMB_1.1.11 memoise_2.0.1
## [184] beeswarm_0.4.0 AnnotationDbi_1.70.0 cluster_2.1.8.1
## [187] globals_0.18.0 GSEABase_1.70.0