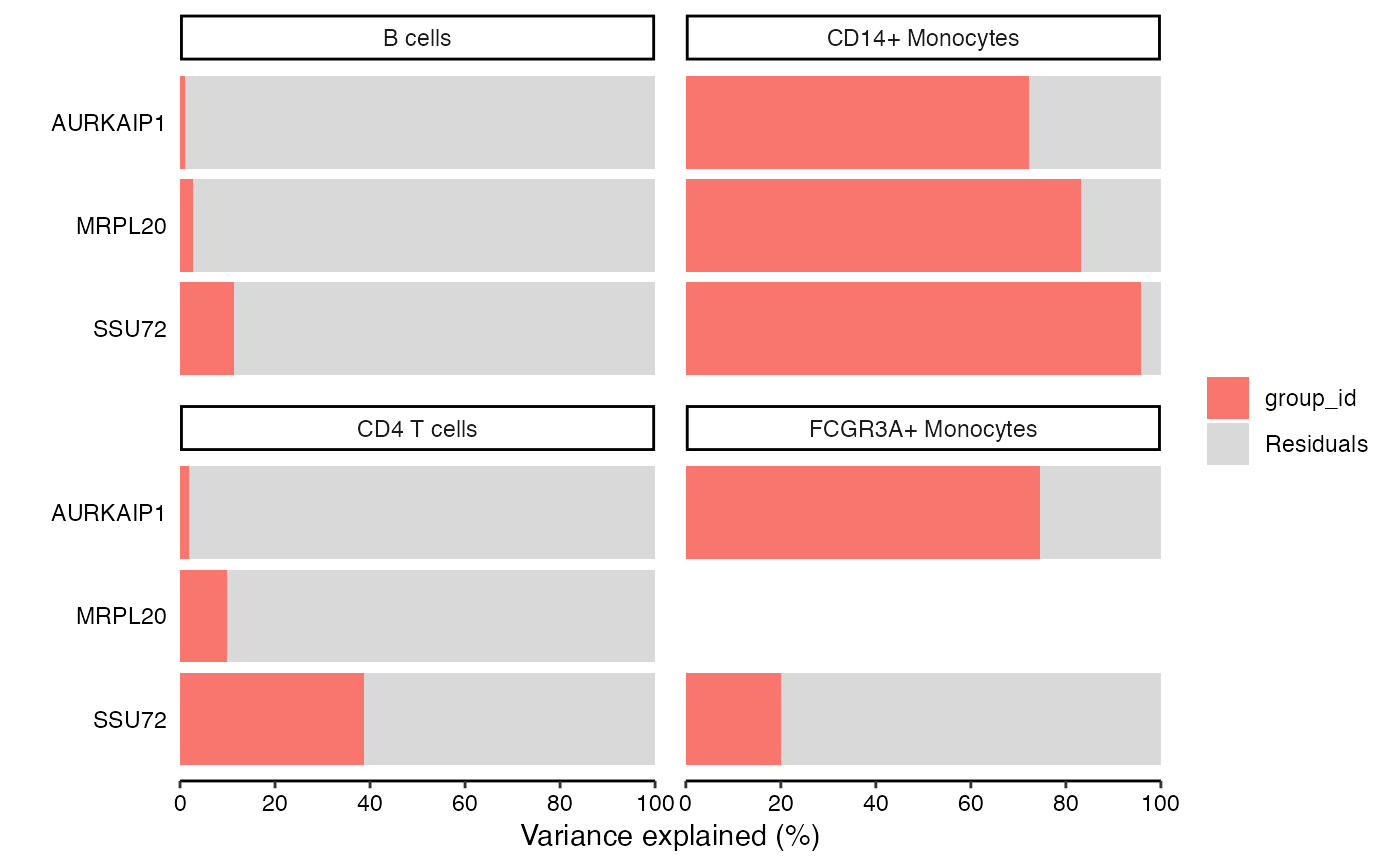
Bar plot of variance fractions for a subset of genes
Arguments
- x
vpDFobject returned byfitVarPart()- col
color of bars for each variable
- genes
name of genes to plot
- width
specify width of bars
- ncol
number of columns in the plot
- ...
other arguments
Examples
library(muscat)
library(SingleCellExperiment)
data(example_sce)
# create pseudobulk for each sample and cell cluster
pb <- aggregateToPseudoBulk(example_sce,
assay = "counts",
cluster_id = "cluster_id",
sample_id = "sample_id",
verbose = FALSE
)
# voom-style normalization
res.proc <- processAssays(pb, ~group_id)
#> B cells...
#> 0.052 secs
#> CD14+ Monocytes...
#> 0.084 secs
#> CD4 T cells...
#> 0.061 secs
#> CD8 T cells...
#> 0.037 secs
#> FCGR3A+ Monocytes...
#> 0.089 secs
# variance partitioning analysis
vp <- fitVarPart(res.proc, ~group_id)
#> B cells...
#> 0.61 secs
#> CD14+ Monocytes...
#> 0.79 secs
#> CD4 T cells...
#> 0.63 secs
#> CD8 T cells...
#> 0.38 secs
#> FCGR3A+ Monocytes...
#> 0.76 secs
#>
# Show variance fractions at the gene-level for each cell type
plotPercentBars(vp, genes = vp$gene[2:4], ncol = 2)
#> Warning: Ignoring empty aesthetic: `width`.