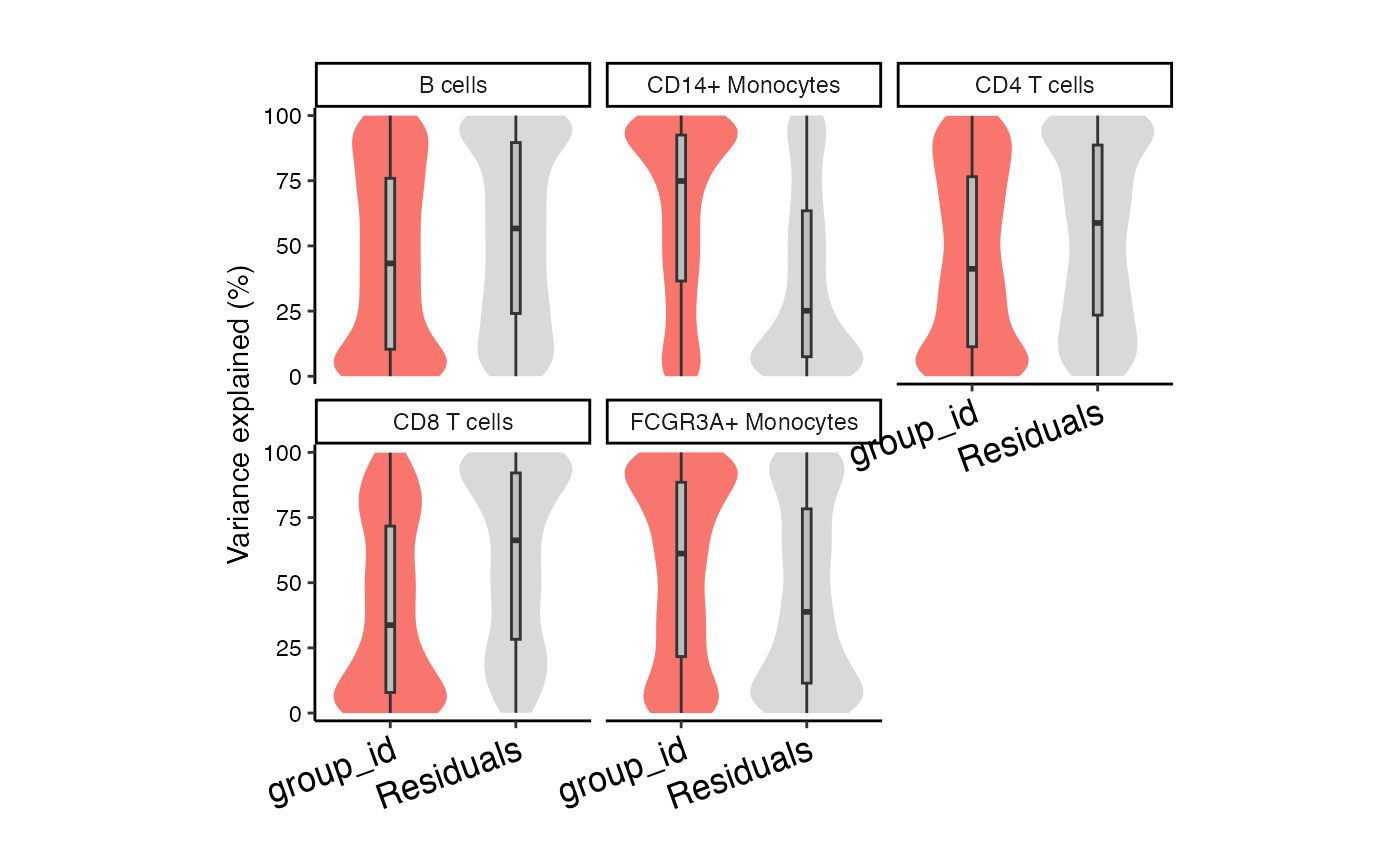
Violin plot of variance fraction for each gene and each variable
Examples
library(muscat)
library(SingleCellExperiment)
data(example_sce)
# create pseudobulk for each sample and cell cluster
pb <- aggregateToPseudoBulk(example_sce,
assay = "counts",
cluster_id = "cluster_id",
sample_id = "sample_id",
verbose = FALSE
)
# voom-style normalization
res.proc <- processAssays(pb, ~group_id)
#> B cells...
#> 0.053 secs
#> CD14+ Monocytes...
#> 0.085 secs
#> CD4 T cells...
#> 0.062 secs
#> CD8 T cells...
#> 0.039 secs
#> FCGR3A+ Monocytes...
#> 0.092 secs
# variance partitioning analysis
vp <- fitVarPart(res.proc, ~group_id)
#> B cells...
#> 0.6 secs
#> CD14+ Monocytes...
#> 0.83 secs
#> CD4 T cells...
#> 0.64 secs
#> CD8 T cells...
#> 0.38 secs
#> FCGR3A+ Monocytes...
#> 0.78 secs
#>
# Summarize variance fractions genome-wide for each cell type
plotVarPart(vp)