Sort variance partition statistics
Usage
# S4 method for class 'vpDF'
sortCols(
x,
FUN = sum,
decreasing = TRUE,
last = c("Residuals", "Measurement.error"),
...
)Arguments
- x
object returned by
fitVarPart()- FUN
function giving summary statistic to sort by. Defaults to sum
- decreasing
logical. Should the sorting be increasing or decreasing?
- last
columns to be placed on the right, regardless of values in these columns
- ...
other arguments to sort
Examples
library(muscat)
library(SingleCellExperiment)
data(example_sce)
# create pseudobulk for each sample and cell cluster
pb <- aggregateToPseudoBulk(example_sce,
assay = "counts",
cluster_id = "cluster_id",
sample_id = "sample_id",
verbose = FALSE
)
# voom-style normalization
res.proc <- processAssays(pb, ~group_id)
#> B cells...
#> 0.061 secs
#> CD14+ Monocytes...
#> 0.084 secs
#> CD4 T cells...
#> 0.061 secs
#> CD8 T cells...
#> 0.037 secs
#> FCGR3A+ Monocytes...
#> 0.079 secs
# variance partitioning analysis
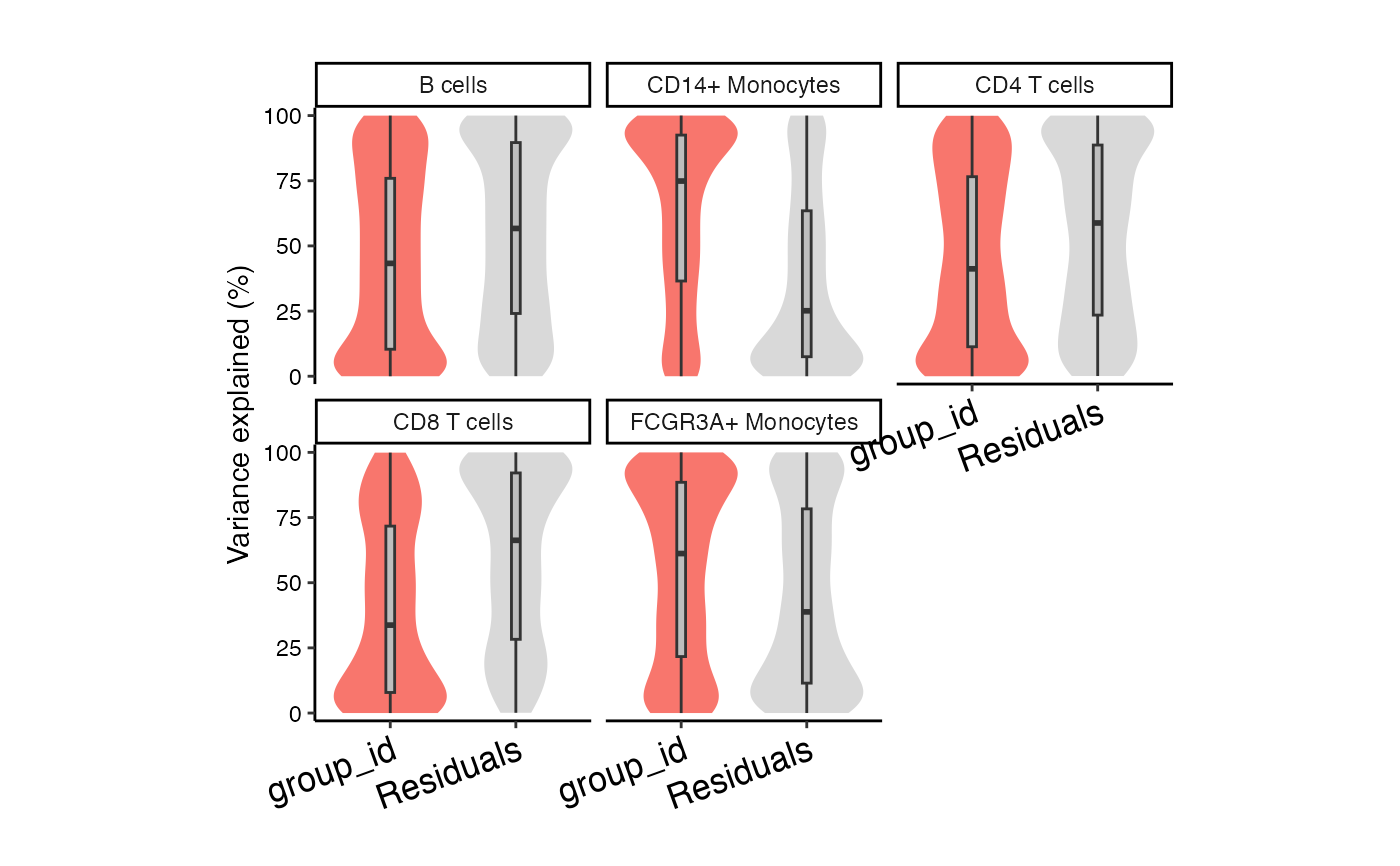
vp <- fitVarPart(res.proc, ~group_id)
#> B cells...
#> 0.59 secs
#> CD14+ Monocytes...
#> 0.81 secs
#> CD4 T cells...
#> 0.63 secs
#> CD8 T cells...
#> 0.39 secs
#> FCGR3A+ Monocytes...
#> 0.75 secs
#>
# Summarize variance fractions genome-wide for each cell type
plotVarPart(sortCols(vp))